
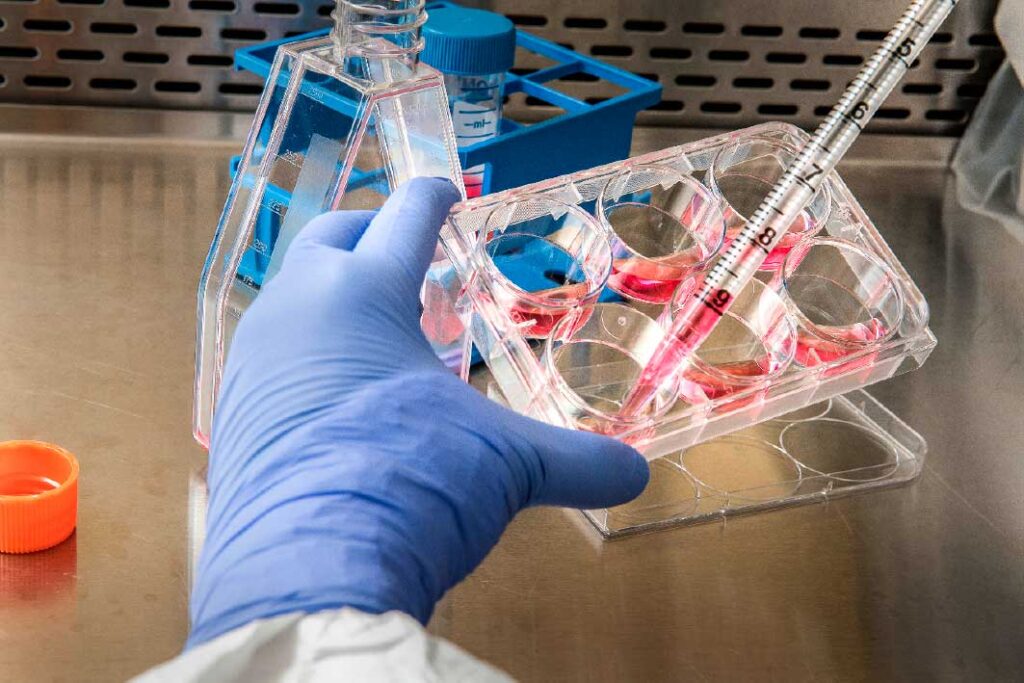
Peroxisome biogenesis disorders Zellweger Spectrum (PBD-ZSD) are rare, complex autosomal disorders affecting the function and formation of peroxisomes.

Treatments for Peroxisome Biogenesis Disorders is only the beginning.
The work being done by PBD Project is going to help millions of people.
Our mission
Our mission is to research and fund innovative therapies,as well as treatment options, expert care, and information to families affected by this disease. We believe in the power of collaboration and communication and our goal is to establish a network of knowledge that will push research forward to find therapies for PBD patients.

PBD Project has had more than one birth.
PBD Project was first born on October 2017, even if we were unaware of it at the time. That is the day Diego was born. We counted 10 fingers, we checked his breath. Everything seemed to be in order.
PBD Project was then born in the summer of 2019, when we first heard “Peroxisome Biogenesis Disorder” for the first time. Little did we know we would end up being armchair experts on PBDs and PEX10.
PBD Project is also born today and tomorrow, and every time it reaches a new parent, new medical expert and a new research milestone.
By bringing together exceptional talent from around the world, we will find a more efficient approach that will eventually lead to scientific advancements and ultimately a cure.
We as parents, will do anything to provide Diego, and patients like him, with the opportunity to a life they deserve.
Scientific Collaborators
Core Team
Tania Rodríguez Sales
With a Bachelor of Arts in International Relations from Instituto Tecnológico y de Estudios Superiores de Monterrey in Mexico City and a specialization in Latin American Politics from Pontificia Universidad Católica de Chile in Santiago (2000). Tania brings over 23 years of dynamic experience in public relations and strategic communications. Passionate about writing and content generation, Tania excels in campaign creative development, audience engagement, strategic messaging, social media strategy, event planning, and budgeting. Their expertise makes her a versatile and driven professional in the field.Jorge Ramón Castillo Soriano
Designer and artist from Mexico City with 15 years of experience in advertising and brand development, JRCS brings creativity, visual complexity and digital skills to PBD Project, he has worked in worldwide agencies like Mc Cann Erickson and Teran/TBWA Mexico. Passionate about arts, illustration, graphic and web design.
2021© All rights reserved by PBD Project
